Top 5 Myths in HPC for Life Sciences
At WekaIO, we have a huge amount of experience in a number of High-Performance Computing areas, ranging from algorithmic trading and financial technology to media and entertainment, oil and gas, AI/ML acceleration, and life sciences. Over the years we’ve discovered a number of traits within life sciences research that can be addressed with a good technical architecture to overcome business challenges (and in some cases business issues that no architecture can ever overcome…but that’s a blog for a different day…). Some of the larger challenges that life sciences companies deal with vary across the work being done, whether it’s drug discovery, genomic sequencing, or other cellular analysis work.
As Weka has discovered, there is a common thread of key tenets that need to be addressed overall: The need for faster result times for any analysis being done, the sprawl of data at scale, the creation of simplified workflows to speed up researcher productivity, and more. So without further ado, let’s take a look at some of these challenges and the myths associated with them.
Myth #1: Data workflows in life sciences are all throughput-sensitive.
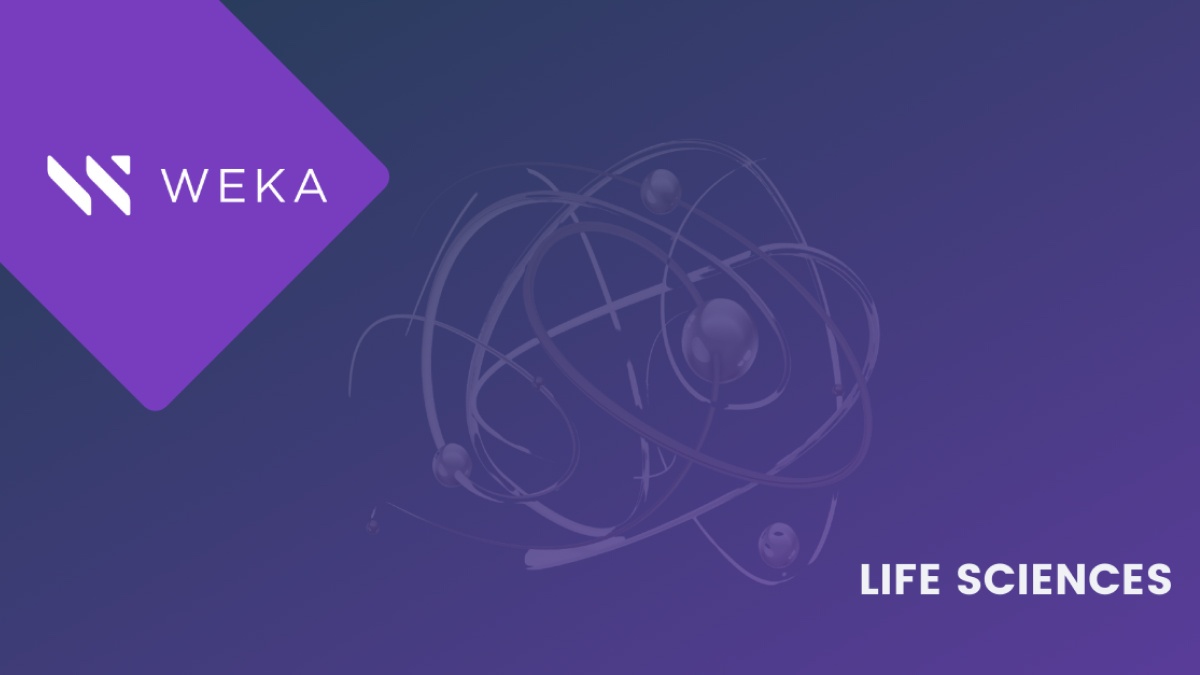
Our Mythbusting Wek-A-meter™ says that, with few exceptions, this myth is false. In fact, we need to segment this category into two different areas: single application usage and at-scale parallel workflows. In single application usage, you may find that you have a self-contained application with dedicated resources for ingest, analysis tools, and output, and some of these very specialized applications will be throughput sensitive only on their own. However, at scale, the combination of many ingest devices, parallel analytics streams, integration into AI/ML workflows, and different data science tools working through the data means that there is essentially a heavy mix of very low-latency, small-file, and metadata-driven functions, and throughput. It’s not unusual to see multi-millions of IOPS needed for both read and write at the same time that 10, 20, 30 Gigabytes per second are needed. The storage system that can handle this level of performance well as you reach into the typical hundreds to thousands of devices and applications in modern research labs can accelerate time-to-discovery and increase the productivity of the research team.
Myth #2: All devices and apps speak common protocols, making them easy to integrate together.
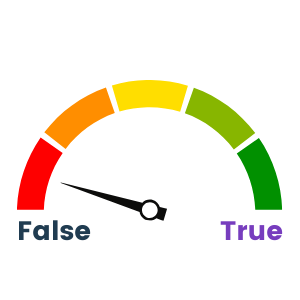
 The Wek-A-Meter™ says that this statement is mostly false. From an infrastructure point of view, most companies try and standardize to be more efficient, but each vendor has preferred methods of communication. Even at the basic data creation layer, you have tools such as cryo-electron microscopes from different manufacturers needing to communicate via SMB for one and via NFS for another. Alternately, certain applications require communication with a POSIX-compliant layer instead. More recently, we’re seeing edge sensors and IoT devices sending data via an S3 API as their native method as well. This adds up to a massive problem with islands of storage for your data depending on the ingest protocol, and even if the data can be centralized on one platform, how do you ensure that it can be accessed concurrently by all applications as needed to create a true data lake?
The Wek-A-Meter™ says that this statement is mostly false. From an infrastructure point of view, most companies try and standardize to be more efficient, but each vendor has preferred methods of communication. Even at the basic data creation layer, you have tools such as cryo-electron microscopes from different manufacturers needing to communicate via SMB for one and via NFS for another. Alternately, certain applications require communication with a POSIX-compliant layer instead. More recently, we’re seeing edge sensors and IoT devices sending data via an S3 API as their native method as well. This adds up to a massive problem with islands of storage for your data depending on the ingest protocol, and even if the data can be centralized on one platform, how do you ensure that it can be accessed concurrently by all applications as needed to create a true data lake?
Top Tip: To solve this issue, Weka storage can handle data intake from any application or device AND present the same data on any of the standardized protocols (POSIX, NFS, SMB, S3, GPUDirect ® Storage). With most other storage platforms, the workflow slows down by copying data and make it accessible to the apps that need it, even within the same storage system
Myth #3: All data flows in life sciences are similar.
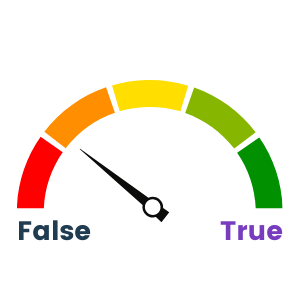
 Wek-A-Meter™ calls this one as 50/50. There have been some strong efforts to try and pull data flows together using things like R-project for statistical analysis, and there are life-science-specific common libraries for AI/ML that are being developed to create commonality for certain groups or classes of data, but it still has a long way to go. The issues I spoke of in Myth #2 still apply, whereby not having truly universal access to each dataset in the data lake is a barrier to standardizing data flows. As a result, we see groupings of data flows and then copying out to different applications to do Extract/Transform/Load (ETL) to provide data into secondary data flows. Because many of the functions in the life sciences space are used widely (PLM, discovery, clinical trial results, compliance, etc.) and from many of the software vendors (Bioclinica, Microsoft, SAS, SAP, etc.) along with widely shared R-based code, there is a modicum of commonality, but not for all of it.
Wek-A-Meter™ calls this one as 50/50. There have been some strong efforts to try and pull data flows together using things like R-project for statistical analysis, and there are life-science-specific common libraries for AI/ML that are being developed to create commonality for certain groups or classes of data, but it still has a long way to go. The issues I spoke of in Myth #2 still apply, whereby not having truly universal access to each dataset in the data lake is a barrier to standardizing data flows. As a result, we see groupings of data flows and then copying out to different applications to do Extract/Transform/Load (ETL) to provide data into secondary data flows. Because many of the functions in the life sciences space are used widely (PLM, discovery, clinical trial results, compliance, etc.) and from many of the software vendors (Bioclinica, Microsoft, SAS, SAP, etc.) along with widely shared R-based code, there is a modicum of commonality, but not for all of it.
Myth #4: Technology can solve all of the issues in life sciences workflows.
Nope. Not even close. Life sciences share one characteristic with most enterprise companies: departmental segregation of resources and budgets. Because of this, only forward-thinking CxO’s/Directors/Business line owners who decide to create sharable resources, both across a company and within departments, will be able to highly optimize their productivity. Sometimes it takes policy wonks and finance gurus to make the needs of the researchers happen effectively.
Myth #5: “Big Data” is larger than you think.
 The Wek-A-Meter™ Is almost off the charts true on this. When Weka surveyed our customers and prospects we discovered that most of them struggled to quantify both how much data they had and the rate of data increase across their environments. Even worse, most struggled to determine which data was active: “hot” vs. “cool” vs. “cold.” As a result, across the islands of storage, it wasn’t unusual to see a 1.5x-2x disparity in the amount of data the customers had compared to what they thought they had. The growth rates in genomics and life sciences are enormous as well. Many of our life sciences customers have centralized tens of petabytes on Weka and continue to grow at petabyte per-month speed. One of Weka’s customers, Genomics England thought they had 20 petabytes of data, but as they centralized and migrated all of their sequencing to WekaFS, they discovered that they actually had close to 40 petabytes. In fact, after they were on Weka they were able to more accurately monitor their growth rate and are now planning for 140 Petabytes of genomics record data by 2023. This story is more common than you may think in life sciences even though the scale may vary.
The Wek-A-Meter™ Is almost off the charts true on this. When Weka surveyed our customers and prospects we discovered that most of them struggled to quantify both how much data they had and the rate of data increase across their environments. Even worse, most struggled to determine which data was active: “hot” vs. “cool” vs. “cold.” As a result, across the islands of storage, it wasn’t unusual to see a 1.5x-2x disparity in the amount of data the customers had compared to what they thought they had. The growth rates in genomics and life sciences are enormous as well. Many of our life sciences customers have centralized tens of petabytes on Weka and continue to grow at petabyte per-month speed. One of Weka’s customers, Genomics England thought they had 20 petabytes of data, but as they centralized and migrated all of their sequencing to WekaFS, they discovered that they actually had close to 40 petabytes. In fact, after they were on Weka they were able to more accurately monitor their growth rate and are now planning for 140 Petabytes of genomics record data by 2023. This story is more common than you may think in life sciences even though the scale may vary.
Top Tip: WekaFS can scale to multiple exabytes in a SINGLE namespace. This allows for all applications to take advantage of data access without having to copy the data around. In addition, Weka can automatically tier data on a policy-driven basis from “hot“ NVMe high-performance flash down to an object store capacity tier attached to the same namespace. This simplifies identifying what data is hot/cool/cold, allows for dynamic expansion of capacity by increasing the object store bucket attached to the namespace, allows for additional protection of the data using Snap-to-object technology from Weka, and can dramatically improve the economics of storage at scale while maintaining high performance.
Life sciences in all its applications from genomics to clinical trials, drug discovery, healthcare, and more are seeing a revolution where the data environments are increasingly taking on characteristics of high-performance computing. Whether it’s doing high-speed analytics or device data ingestion or managing PLM and clinical trials data, the requirements for capable storage are always growing. A key factor in successful data workflow optimization is having a storage platform that can meet the needs for speed, scale, and simplicity. Weka provides all of these capabilities and more to give you improved time-to-value for your data.

Additional Helpful Resources
WekaFS for Life Sciences: Accelerate the data pipeline in Life Sciences
Modern Workloads in Pharma and Life Sciences
AI-based Drug Discovery with Atomwise and Weka on AWS
Accelerating Cryo-EM & Genomics Workflows
Accelerating Genomic Discovery with Cost-Effective, Scalable Storage
Accelerating Discovery and Improving Patient Outcomes With Next-Generation Storage
How to Analyze Genome Sequence Data on AWS with WekaFS and NVIDIA Clara Parabricks





