How to Accelerate Drug Discovery with Cryo-EM in the Cloud
Cryo-EM pipelines create significant computing and data storage challenges that can be efficiently and cost-efficiently addressed in the public cloud
Understanding protein structure and the physical arrangement of amino acids at binding sites plays a vital role in drug discovery, enabling scientists to design and develop targeted drugs selectively and effectively.
A major limitation in structure-based drug discovery is the availability of ‘experimentally determined high resolution structures.’ X-ray crystallography has been relied on to solve protein structures in the past, but it can be a very laborious process. Because of experimental limitations, not all protein structures can be determined using X-Ray crystallography.
Cryogenic electron microscopy (Cryo-EM) has emerged as an important structural discovery tool, creating a new approach for solving protein structures at high resolution quickly and accurately that complements other methods such as X-Ray crystallography and AI-based structural prediction.
Using Cryo-EM, scientists capture thousands of high resolution 2D images of flash-frozen protein molecules in solution. Specialized image processing software utilizing High Performance Computing (HPC) then transforms these images into dynamic 3D models with near-atomic resolution. A single Cryo-EM run may generate as much as 10TB of raw data.
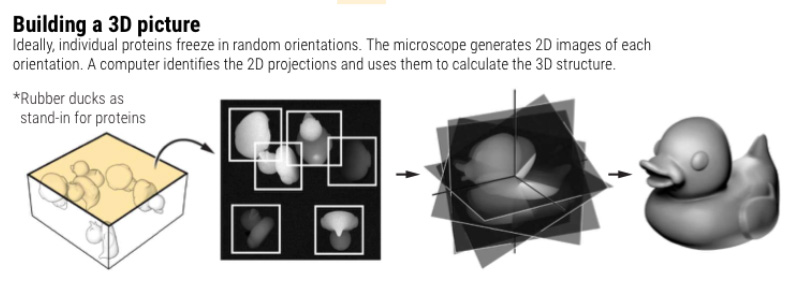
However, the large-scale data generated by Cryo-EM presents a significant data storage and computing challenge. Timely processing of Cryo-EM data is essential to identify new potential drugs. Massive GPU and CPU capacity along with high-performance data storage are needed to quickly convert raw Cryo-EM data into accurate structural models. Cryo-EM computing environments must be able to:
- Deliver results fast: Return protein structures quickly, enabling drug design and discovery to proceed.
- Simplify deployment and management: Enable teams to focus on science, not HPC infrastructure.
In addition, many implementing Cryo-EM are struggling to gain control over compute, data storage and software license costs. An HPC cluster requires expensive CPU, GPU, network and storage resources, and the license costs for software tools like Cryo-SPARC can be significant. Building an environment that controls costs and ensures maximum utilization and ROI is a challenge.
Cloud computing can mitigate risk and prevent delay by providing unlimited compute (CPU/GPU) and data storage on-demand to process Cryo-EM data and predict structures quickly, enabling large-scale screening of chemical compound libraries to find novel drugs.
Cryo-EM computing and data challenges
Cryo-EM data processing transforms raw 2D image data into 3D models and motion clips using specialized software such as RELION and CryoSPARC. This software performs a variety of computationally intensive tasks such as blur removal, motion correction, and 2D and 3D image classification, creating significant compute and data storage challenges in the process:
Cryo-EM Challenges
- Compute: Cryo-EM pipelines have been adapted to take advantage of the latest GPUs as well as newer CPU instruction sets such as AVX-512. It can be difficult to feed data to high-performance computing hardware fast enough to ensure high utilization.
- Data storage: The steps in the Cryo-EM data processing pipeline have a high degree of variability. Some steps require fast sequential access while others require random access. In practice, this variability often leads to time-wasting data copies to local storage between each step—or reduced performance for steps where storage performance is suboptimal.
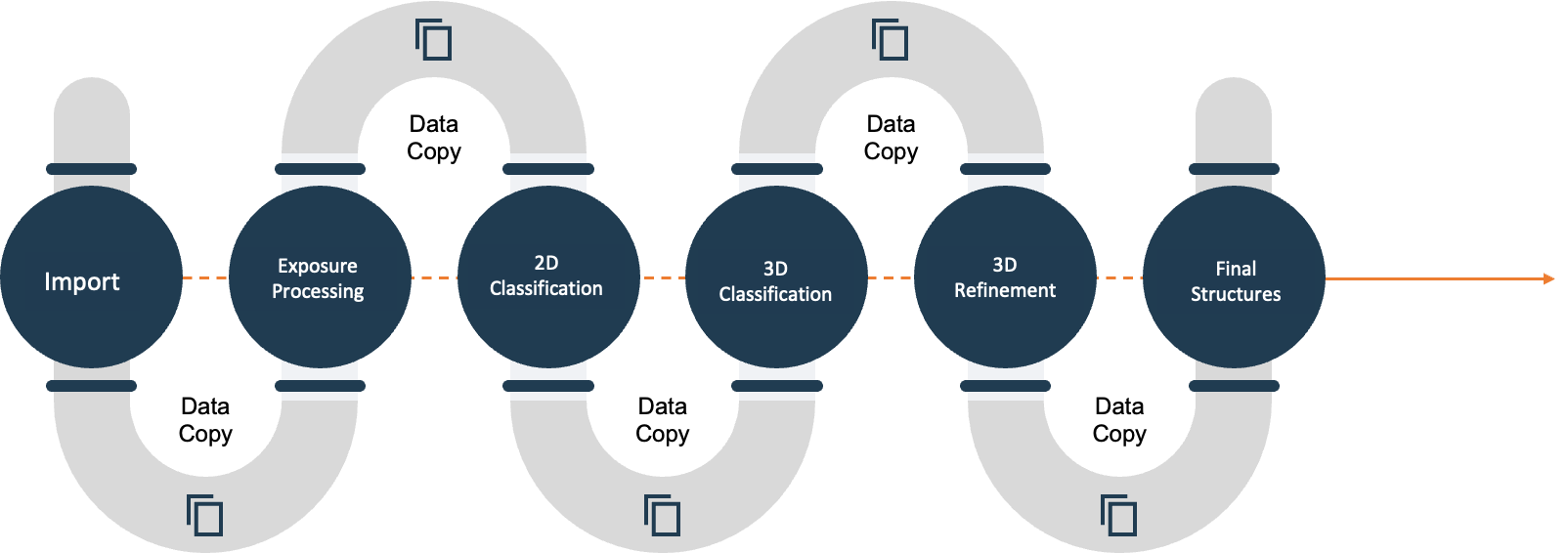
Cryo-EM’s compute and storage needs create significant challenges for researchers and IT teams:
- How do you keep computing and data I/O from becoming bottlenecks to Cryo-EM data processing?
- How do you ensure that you’re maximizing utilization of expensive compute, storage and software resources?
- How do you build an optimized HPC cluster for Cryo-EM data processing?
- How do you get access to the latest GPUs, CPUs, and high performance storage?
- How do you keep up with rapid technology evolution?
How can the cloud accelerate Cryo-EM workflows?
Many Cryo-EM labs are turning to the cloud to help solve these challenges because they can start to deploy infrastructure in a more on-demand fashion, they can improve their ability to elastically scale their infrastructure, and they can gain continual access to the latest and greatest infrastructure offerings from their cloud provider.
- Improved agility: The ability to spin up an entire infrastructure stack in just a few minutes, enables labs to stand up a new Cryo-EM environment in just a few minutes and dramatically improve time to results. Long procurement and hardware deployment cycles become a thing of the past.
- Elastic scaling: Because labs can scale resources up to meet project timelines and then scale the entire project back down, labs don’t need to over-provision resources or pay for resources they don’t actually use.
- On-demand access to infrastructure advances: In recent years, cloud providers have made big leaps providing access to high performance infrastructure options like GPU-enabled compute machines and 400 GbE networking. The pace of innovation in cloud infrastructure shows no signs of slowing. As a result, labs have the ability to continually access faster networks and high performance compute capabilities without waiting for hardware refresh cycles.
This article has been co-developed with our WEKA X partner, Clovertex, a cloud organization specializing in architecting, automating, and managing applications for HPC in the cloud. Clovertex provides solutions tailored to specific research needs that allow HPC workloads to move seamlessly to the cloud.


